• General Information
• Universal TERMs
• Matches & Database
• Extracting Info
• Relevant References
General Information
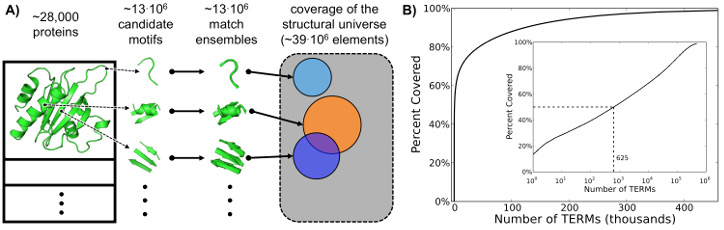
To arrive at a list of universal TERMs, we undertook the task of finding the smallest subset of all possible TERMs, each defined to capture the full structural environmnent around a single residue in our structural database, that maximally describes the structural information within the database [1]. Here we deposit the results of this analysis — the universal TERMs themselves (as fragments in PDB format) and all their matches, as defined in the paper. From these data, it is possible to generate sequence alignments corresponding to each TERM, generate the structures of all the matches for any given TERM, and other analyses.
Getting Universal TERMs
We have deposited all of the relevant data from our universal TERM analysis in a public rsync module on the server grigoryanlab.org. The relevant parts to download depend on your intended application. But in all cases, start with downloading the file coverage.txt, which shows how fraction of the structural universe described (from 0 to 1) increases as a function of the number of TERMs considered. To download this file, run the command:
rsync -vaRrz grigoryanlab.org::univ-TERMs/coverage.txt /local/path/
where /local/path is the local path where you want downloaded files to reside. If you take a look inside this file, you will find something like this:
% head coverage.txt
0 0.134229351408
1 0.194978515915
2 0.225194452488
3 0.239854346516
4 0.253062793009
5 0.263250866799
6 0.270368007889
7 0.276267013125
8 0.280901392357
9 0.285289467711
If you want to download all TERMs we have compiled, use the following command (note, this means over 450,000 TERMs; in most cases, you will want a subset; see info on subsets below):
rsync -vaRrz grigoryanlab.org::univ-TERMs/TERMs /local/path/
This will create the directory /local/path/TERMs with sub-directories that will contained individual PDB files, one for each motif. The PDB files are named by TERM priority (starting from zero) — e.g., file /local/path/TERMs/000/000125.pdb corresponds to the 126-th motif, in the order discovered by the set cover procedure [1]. This can be loosely interpreted as the 126-th most "important" motif for describing the structural universe.The hierarchical layout of this directory is to deal with the large total number of motifs (we include all motifs to go up ~99% of coverage). The sub-directory name within the root TERMs directory derives from the first three digits of the TERM number, when written up to the hundred-thousaunds place in decimal notation. E.g., the 1,235-th TERM is in file TERMs/001/001234.pdb and the 12,347-th TERM is in TERMs/012/012346.pdb.
TERM Subsets
If you don't necessarily need all universal TERMs, it is quite easy to download just the subset you want. This is because TERMs/000 contains the first 1000 TERMs (files 000000.pdb to 000999.pdb). So, if you want to download no more than the first 1000 TERMs, you would simply run:
rsync -vaRrz grigoryanlab.org::univ-TERMs/TERMs/000 /local/path/
Similarly, if you want to download just the top 10000 TERMs, simply run:rsync -vaRrz grigoryanlab.org::univ-TERMs/TERMs/00* /local/path/
Note, that this will finish must faster than downloading the entire database.Matches and the Database
You have downloaded the TERMs you wanted and now you want to see which proteins they occur in and with what sequences. You can, of course, simply use our MASTER search engine to search for instances of any given TERM in whatever structural database you like. But if you want to get the precise matches and sequences that we identified in the 2016 PNAS paper [1], you can do so by downloading the PDS file corresponding to each TERM (a processed version of the PDB file, that can be used as input into MASTER), the match file corresponding to each TERM (which lists all matches for the TERM, as defined in the paper), and the search database. Download the search database by running the following:
rsync -vaRrz grigoryanlab.org::univ-TERMs/db /local/path/
Next, as with TERMs themselves, you can either download a subset of PDS and match files (corresponding to the subset of TERMs you downloaded above) or, if you really-really want, you can download all PDS and match files (for all TERMs); again, the latter will take a while. Assuming you want a subset, you would do something like:
rsync -vaRrz grigoryanlab.org::univ-TERMs/TERMs-pds/00* /local/path/
rsync -vaRrz grigoryanlab.org::univ-TERMs/matches/00* /local/path/
rsync -vaRrz grigoryanlab.org::univ-TERMs/TERMs-pds /local/path/
rsync -vaRrz grigoryanlab.org::univ-TERMs/matches /local/path/
Adjusting match files
Match file store the exact specification for a particular match to a TERM, one match per line, including the path to the corresponding database entry. Because you can place your downloaded database under an arbitrary path, you need to adjust all match files by pre-pending the correct global path prefix to the database file identifier. For example, a particular match file may look like:
% head -3 matches/001/001035.match
2.61174e-07 vt/3vtu [(58,62), (66,70)]
0.181367 me/4me9 [(294,298), (302,306)]
0.198307 wa/3wan [(188,192), (196,200)]
% head -3 matches/001/001035.match
2.61174e-07 /local/path/db/vt/3vtu.pds [(58,62), (66,70)]
0.181367 /local/path/db/me/4me9.pds [(294,298), (302,306)]
0.198307 /local/path/db/wa/3wan.pds [(188,192), (196,200)]
for i in path-to-matches/*; do echo $i; for j in $i/*; do awk -F' ' -vOFS=' ' '{ $2 = "path-to-db/"$2".pds" " " }1' $j > $j.tmp; mv $j.tmp $j; done; done
where you should replace path-to-matches with the path to your matches directory and path-to-db with the path to your downloaded search database (i.e., /local/path/db in the example above).Extracting Matches and Sequences
Once all of the above operations are performed, you can extract structural and sequence information for any TERM's maches using MASER. Suppose you want to analyze the 582-nd TERM. The corresponding match file, matches/000/000581.match , contains 3952 lines, meaning that we found 3592 matches to the TERM in our analysis. To dump the sequences of all matches, you could say:
/master-path/master --query /local/path/000/000581.pds --matchIn /local/path/matches/000/000581.match --seqOut /out/path/000581.seq
where /master-path is a directory where MASTER binaries reside. The file /out/path/000581.seq will then contain the sequence of every match, in the same order as listed in the match file. If the structure of every match is also desired, the following command can be used:/master-path/master --query /local/path/000/000581.pds --matchIn /local/path/matches/000/000581.match --seqOut /out/path/000581.seq --structOut /out/path/000581.struct
in which case a directory called /out/path/000581.struct will be created and match PDB files will be written into it, one file per match, with names corresponding to match number. MASTER offers additional output options (e.g., one can output full structures, oriented onto the query structure via just the matching region), see here for more information.Relevant References
C. O. Mackenzie, J. Zhou, G. Grigoryan, "Tertiary Alphabet for the Observable Protein Structural Universe", Proceedings of the National Academy of Sciences, 113(47): E7438-E7447, 2016.
J. Zhou, G. Grigoryan, "Rapid Search for Tertiary Fragments Reveals Protein Sequence-Structure Relationships", Protein Science, 24(4): 508-524, 2015.
F. Zheng, J. Zhang, G. Grigoryan, "Tertiary Structural Propensities Reveal Fundamental Sequence/Structure Relationships", Structure, 23(5): 961-971, 2015.